
I am a Fulbright alumni coming from Bulgaria, currently a bioinformatics postdoc in UC Berkeley.
Received my bachelor's degree from Sofia University "St. Kliment Ohridski" in molecular biology and my masters and PhD from Georgia Institute of Technology in bioinformatics. I have spent one year in the Netherlands following
a masters degree in Molecular Life Sciences. After that I took the opportunity to study in the USA through a Fulbright scholarship.
I got interested in bioinformatics because I noticed a certain cycle of information between the wet lab and the dry lab, proving that
it is important to know equally well both fields.
11. Penev P.I., Alvarez-Carreño C., Smith E., Petrov A.S.,Williams L.D. "TwinCons: Conservation score for uncovering deep sequence similarity and divergence", PLoS Comp Biol (2021)
10. Penev P.I., McCann H.M., Meade C.D., Maddala A., Bernier C.R., Chivukula V.L., Ahmad M., Gulen B., Sharma A., Alvarez-Carreño C., Williams L.D., Petrov A.S. "ProteoVision: web server for advanced visualization of ribosomal proteins", NAR (2021)
9. Penev P.I., Fakhretaha-Aval S., Patel V. J., Cannone J.J., Gutell R.R., Petrov A.S., Williams L.D., Glass J. B. "Supersized ribosomal RNA expansion segments in Asgard archaea", GBE (2020)
8. Lin-Xing Chen, Jaffe A. L., Borges A. L., Penev P.I., Nelson T. C., Warren L. A., Banfield J. F. "Phage-encoded ribosomal protein S21 expression is linked to late stage phage replication", bioRxiv (2021)
7. Alvarez-Carreño C., Penev P.I., Petrov A.S., Williams L.D. "Fold Evolution before LUCA: Common Ancestry of SH3 Domains and OB Domains", MBE (2021)
6. Mascuch S.J., Fakhretaha-Aval S., Bowman J.C., Minh Thu H.Ma, Thomas G., Bommarius B., Ito C., Zhao L., Newnam G.P., Matange K.R., Thapa H.R., Barlow B., Donegan R.K., Nguyen N.A., Saccuzzo E.G., Obianyor C.T., Karunakaran S.C., Pollet P., Rothschild-Mancinelli B., Mestre-Fos S., Guth-Metzler R., Bryksin A.V., Petrov A.S., Hazell M., Ibberson C.B., Penev P.I., Mannino R.G., Lam W.A., Garcia A.J., Kubanek J., Agarwal V., Hud N.V., Glass J.B., Williams L.D., Lieberman R.L. "A blueprint for academic laboratories to produce SARS-CoV-2 quantitative RT-PCR test kits", JBC (2020)
5. Bowman J.C., Petrov A.S., Frenkel-Pinter M., Penev P.I., Williams L.D. "Root of the Tree: The Significance, Evolution, and Origins of the Ribosome", Chem. Rev. (2020)
4. Mestre-Fos S., Penev P.I., Richards J.C., Dean W.L., Gray R.D., Chaires J.B., Williams L.D. "Profusion of G-quadruplexes on both subunits of metazoan ribosomes", PLoS ONE (2019)
3. Mestre-Fos S., Penev P.I., Suttapitugsakul S., Hu M., Ito C., Petrov A.S., Wartell R.M., Wu R., Williams L.D. "G-Quadruplexes in Human Ribosomal RNA", J. Mol. Biol. (2019)
2. Kovacs, N.A., Penev, P.I., Venapally, A., Petrov, A.S., and Williams, L.D. "Circular Permutation Obscures Universality of a Ribosomal Protein", J. Mol. Evol. 581-592 (2018)
1. Bernier, C.R., Petrov, A.S., Kovacs, N.A., Penev, P.I., and Williams, L.D. "Translation: The Universal Structural Core of Life", Mol. Biol. Evol. 35, 2065–2076 (2018)
Currently I am working in professor Banfield's lab
on informational transfer systems in phages and extrachromosomal elements, as well as metagenomic analysis of soil microbial dynamics, combined with time- and depth-dependent variables. I have strong interest in ribosomal evolution and its role in cellular informational transfer.
My PhD dissertation was based on research on ribosomal evolution, the development of computational tools to study sequence alignments, and the development of a web server for advanced visualization of ribosomal proteins.
My github repository contains code, used in various papers and ongoing projects.
During my masters I worked in professor Gaucher's lab on an experimental phylogenetics project. I compared the viability of different
predictive phylogenetic methods in the structural background of proteins.
In the Netherlands I worked in professor Dolf Weijers' Plant Development biochemistry laboratory.
We worked on new possible interactors of the Auxin Response Factor in Arabidopsis Thaliana.
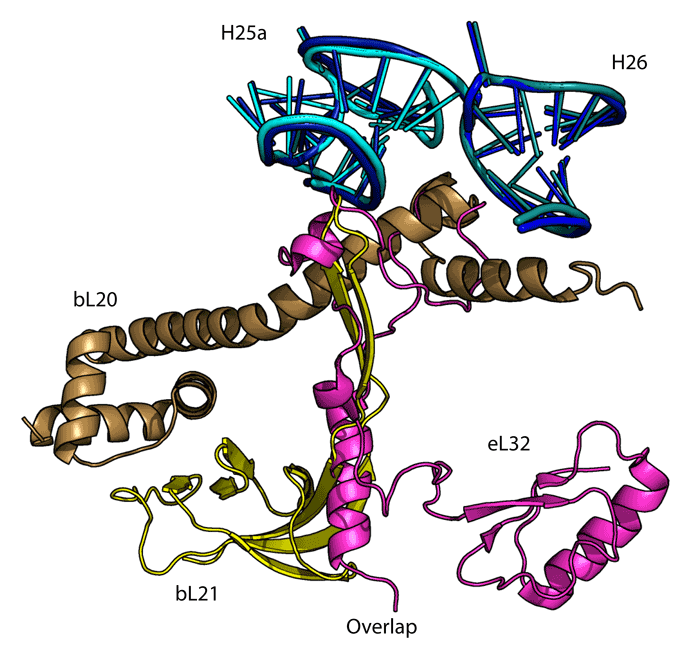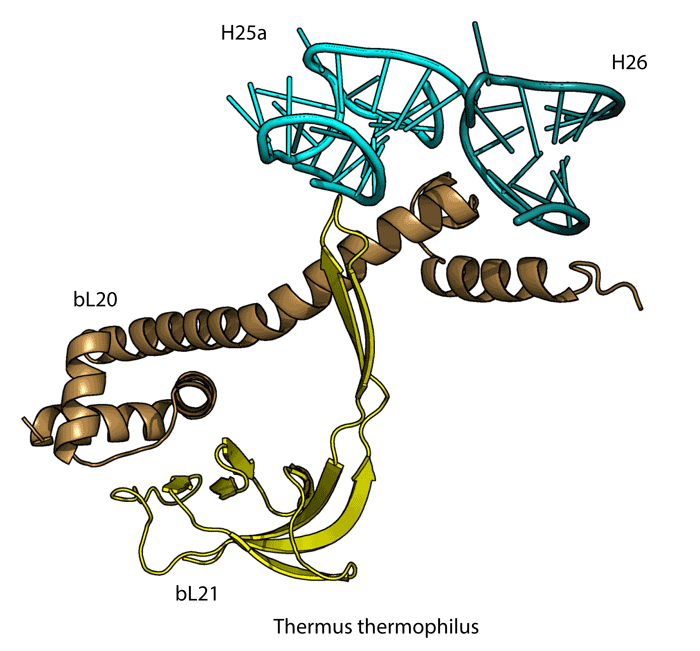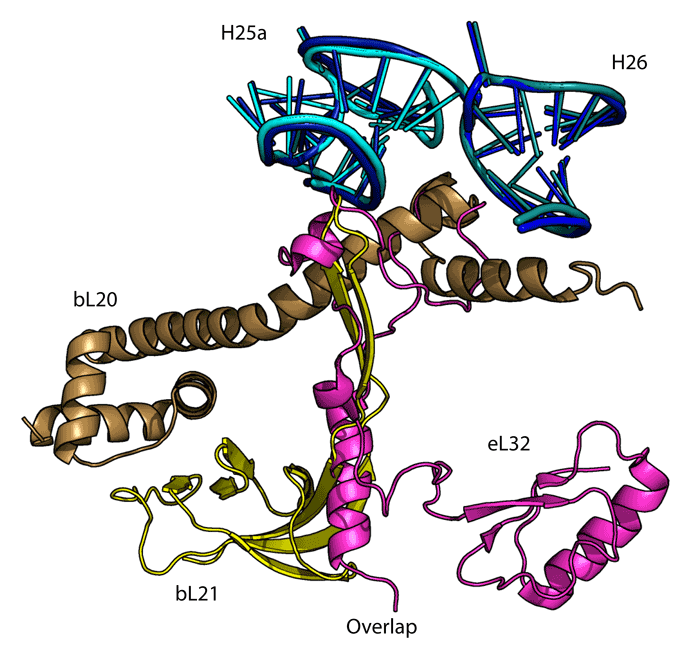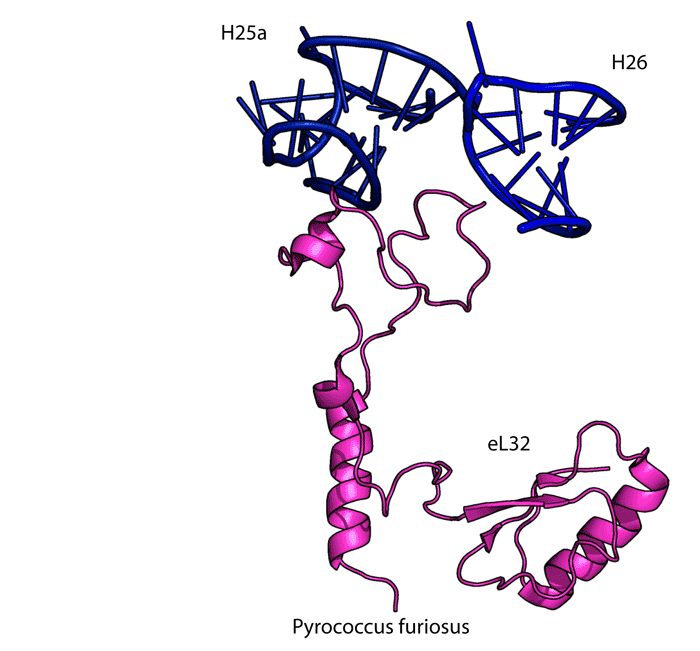
2151 Berkeley Way, Berkeley, CA 94720