FLORA Alignment

Reference
FLORA is being provided on behalf of Susan A. Gerbi. The publication is available here , and the Supplementary Information is available here.
Doris SM, Smith DR, Beamesderfer JN, Raphael BJ, Nathanson JA and Gerbi SA. (2015).
Universal and Domain-Specific Sequences in 23S-28S Ribosomal RNA Identified by Computational Phylogenetics.
RNA 21: 1719-1730.
PMID: 26283689; PMCID: PMC4574749. [
Citation Manager]
Introduction
Welcome to the Full-Length Ribosomal RNA Organismal Alignment (FLORA) database- a global large subunit sequence alignment with broad phylogenetic representation from each domain of life (Bacteria, Archaea, Eukarya) curated from the source database SILVA (Doris et al, 2015). The 23S-28S rRNA sequences in FLORA are full-length, aligned according to secondary structure, and contain a single entry per species. This refinement produced a library of large ribosomal subunit sequences containing 342 Eukaryotes, 915 Bacteria and 86 Archaea (figure below).
Figure
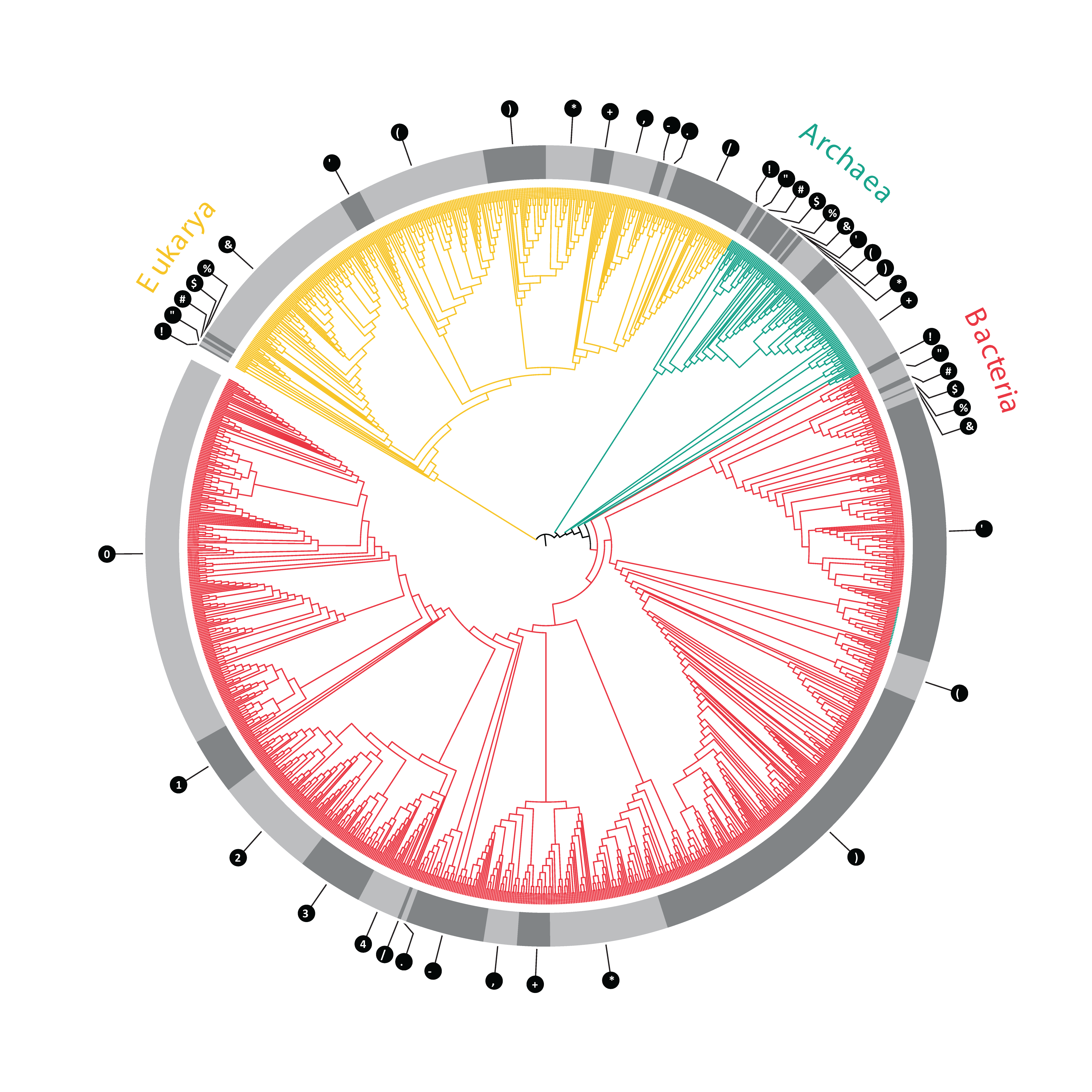
Evolutionary distribution of eukaryotic ( yellow), archaeal ( green), and bacterial ( red) organisms. The phylogenetic relationships of organisms whose 23S-28S rRNA sequences were included in FLORA. The cladogram was derived from the ARB/SILVA LSU Ref guide tree using the interactive tree of life (iTOL) software platform. The number of species included for each group is indicated by the number in the table, below.
|
Eukarya |
Archaea |
Bacteria |
||||||
|
A |
Diplomonads |
2 |
Methanobacteria |
3 |
Aquificae |
4 |
||
|
B |
Parabasalids |
2 |
Methanococci |
5 |
Thermotogae |
10 |
||
|
C |
Amoebozoa |
2 |
Korarchaeota |
1 |
Deinococcus-Thermus |
3 |
||
|
D |
Rhizaria |
1 |
Thermoprotei |
14 |
Chloroflexi |
4 |
||
|
E |
Rhodophyta |
3 |
Methanopyri |
1 |
Dictyoglomi |
1 |
||
|
F |
Embryophyta |
100 |
Thermococci |
5 |
Fusobacteria |
5 |
||
|
G |
Porifera |
12 |
Archaeoglobi |
1 |
Firmicutes |
145 |
||
|
H |
Fungi |
70 |
Thermoplasmata |
3 |
Tenericutes |
22 |
||
|
I |
Alveolata |
34 |
Thermoprotei |
15 |
Cyanobacteria |
186 |
||
|
J |
Stramenophiles |
26 |
Halobacteria |
11 |
Actinobacteria |
64 |
||
|
K |
Mollusca |
11 |
Methanomicrobia |
34 |
Epsilonbacteria |
18 |
||
|
L |
Platyhelminthes |
24 |
PVC-Planctobacteria |
18 |
||||
|
M |
Chordata |
6 |
Spirochaetes |
43 |
||||
|
N |
Arthropoda |
5 |
Acidobacteria |
3 |
||||
|
O |
Nematoda |
40 |
Synergistetes |
2 |
||||
|
P |
Deltaproteobacteria |
23 |
||||||
|
Q |
Mitochondria |
36 |
||||||
|
R |
Alphaproteobacteria |
89 |
||||||
|
S |
Betaproteobacteria |
56 |
||||||
|
T |
Gammaproteobacteria |
179 |
||||||
FLORA Databases (Alignments)
For each phylogenetic domain, raw unfiltered alignments and two structurally filtered alignments, as well as a merger for the large subunit rRNA from the three domains of life, are available for download. Structural filters produce an alignment wherein all columns are structurally homologous to the filtering organism, insertions are excluded, and deletions are held by gaps. The Universal Merged FLORA merges all the domains of life in one alignment and contains only universal positions.
|
Filtered Alignments |
|||
|
Eukaryotic [ S. cerevisiae] |
table (.txt) | FASTA (.fa) | |
|
Eukaryotic [ A. thaliana] |
table (.txt) | FASTA (.fa) | |
|
Archaeal [ H. marismortui] |
table (.txt) | FASTA (.fa) | |
|
Archaeal [ S. solfataricus] |
table (.txt) | FASTA (.fa) | |
|
Bacterial [ E. coli] |
table (.txt) | FASTA (.fa) | |
|
Bacterial [ C. ramosum] |
table (.txt) | FASTA (.fa) | |
|
Unfiltered Alignments (gzipped) |
|||
|
Eukaryotic |
table (.txt) | FASTA (.fa) | |
|
Archaeal |
table (.txt) | FASTA (.fa) | |
|
Bacterial |
table (.txt) | FASTA (.fa) | |
|
Merged Alignments |
|||
|
Universal |
table (.txt) | FASTA (.fa) | |